Here you will obtain a crash-course in DNA, genetics, and the inner-workings of the cell. In doing so, you will probably question whether it is the result of chance and evolution, or of a supreme mind.
Introduction
I began studying genetics in 2011 after I became aware that the genetic code was actually digital. I quickly became enthralled with the whole cellular system, constantly studying all aspects of it including DNA, the genetic code, and the inner workings of the cell especially enzymes; tiny biological molecular machines at the sub-microscopic level which carry out specific tasks that makes life possible. As a former computer programmer in assembly language, I was able to understand how the genetic code is formatted and processed, as it is comparable, in a way, to the manner CPUs handle machine code.
This is my own attempt to write the fundamentals as I understand them, in my own words, as simply as possible for anyone who’s interested in developing a more than basic understanding of this fascinating subject, which seriously challenges evolutionary theory.
Separating the Medium and the Message
First of all, here’s a very important fact that I found out early on, which not only gave me the proper understanding of how genetics works, but allowed me to dive into it all so much more easily as it created a proper way of thinking. It divides the chemicals from the information which is the foundation to being able to grasp the informational aspect of life.
DNA is nothing more than an information storage molecule.
DNA is the medium, not the message. Maintaining this distinction is imperative for understanding the much more involved aspects described throughout this document.
DNA is not related to the genetic code (and genes, as they are sections of genetic code, and the genome, which is the whole of the genetic code as explained later). DNA can be likened to a hard drive, and the genetic code the data it holds, like programs and pictures. There is no connection between the magnetism pattern on a hard drive and the files it stores, and there is no connection between DNA and the genetic code it stores.
Putting it another way, the paper and ink of a book is like the DNA, and the letters and words formed by the arrangement of the ink is like the genetic code, and chapters in the book are like the genes. Chapters divide the information in the book but not the book physically, like genes divide the information in the DNA but not the DNA physically.
This is important as many people believe that DNA and genes/genetic code are the same things, and that genetics is biological chemical reactions. This isn’t the truth; DNA stores digital data, written in a language, which is read and processed much like microchips read and process machine code in computers.
For the sake of simplicity, this document mainly only concentrates on human DNA.
To help make all of this as understandable as possible, I’ve divided it into four main sections: DNA, Proteins and Enzymes, Genetic code (including Genes and Genomes), and The Cell. This is then followed by analysis and conclusions.
DNA
Deoxyribonucleic acid (DNA) is an information storage molecule. It’s the long twisted strand that everyone is familiar with. Imagine it as a ladder that has become twisted. The sides of the ladder are formed of sugar and phosphates. The rungs of the ladder are formed of pairs of chemical bases called Nucleobases (or just “bases” for short). So each rung is aptly called a Base Pair. The bases are a group of four nitrogen-based molecules, which are: Adenine (A), Thymine (T), Cytosine (C), and Guanine (G). There is a fifth one Uracil (U) which is explained later in The Cell section, and it is used when reading DNA. In genetics, the letters are always used instead of the full names of the bases.
Bases pair together following a pairing rule. A and T can only pair, and C and G can only pair. This rule is caused by the molecular make-up of the base molecules. Therefore if we read one side of the DNA strand, we know what the other side should be.
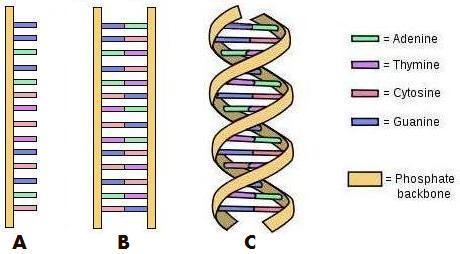
The above 3 drawings will help you understand the construction of DNA. Working from the left, drawing A is a single strand of DNA with single bases. Pause here for one moment - this is an important fact to understand before reading on - the information running from top to bottom in this example is: GGAGACTCTGCGAC (using the first letter of the names of the bases). The different molecules define the code. This is the genetic code which is explained in detail later. To form the complete double-stranded DNA as in B, each base is paired with its complementary base (the only other molecule that it can bond with). The two bases form a bond becoming a base pair, and the “ladder” is complete. This is twisted into the familiar double-helix shape as shown in drawing C.
So you can see that only one strand is required to form the complete double-helix structure, and that either strand can be used to form the other side. This is how DNA is replicated when cells divide. First, the double-helix is “unzipped” into two single strands like in drawing A – the base pairs are pulled apart, straight down the middle. Then, complementary strands are built on each of these strands. New, correct bases are bonded forming base pairs again, finishing with two complete and identical double-helix DNA strands. One stays in the original cell and the other is for the newly replicated cell.
So now you should understand that DNA is “just” the molecule. The important genetic information – genes written in genetic code – is the type and arrangement of the bases.
Note: Each base represents a “bit” of information, like in computers. The difference is that a “bit” in computers is binary (0,1) but a “bit” in DNA can be four values (four bases), so the number system is quaternary (0,1,2,3).
If numbers are assigned to the bases (A=0, C=1, G=2, T=3) then the above DNA example (GGAGACTCTGCGAC) would read 22020131321201.
To finish, here are some interesting facts about the DNA molecule:
- The DNA structure is the densest memory medium known – it’s about 2.5 nanometres wide (40,000 side-by-sides equals the width of a strand of human hair) and is coiled into a ball. If all the letters (A,C,G,T) contained in a pinhead size (2mm diameter) of DNA were written on pages in average sized paperback books, the amount of books would pile to the moon, 500 times. Or in computer terms, 2 million 2-terabyte hard drives worth of data.
- The DNA strand in humans is about 3 billion base pairs long, which makes the actual strand about 6 feet long. But since it’s only 2.5 nanometres wide, it fits in the nucleus of every one of our body’s 10 trillion cells.
- Staying with this – 3 billion quaternary “bits” of information is therefore the size of the human genome (Genome is the term given to the entirety of an organism’s genetic information, encoded in its DNA). 3 billion quaternary bits = 6 billion binary bits = 750 Mbytes if stored on a computer.
Chromosomes
Chromosomes are distinct, separate sections of DNA. An organism’s DNA strand is physically divided into these segments. Since this document is predominantly on the subject of the genetic code, there’s no need to explain chromosomes in any detail here.
Proteins and Enzymes
Before I cover the genetic code, it’s best that I mention what the genetic code is mostly used for – constructing (synthesizing) proteins and enzymes.
Proteins
Proteins are made of a combination of amino acids. There are 20 different amino acids which are all responsible for making a protein a certain way. The amino acids are bonded to each other to form a chain which is then folded into a 3-dimensional structure. They form parts of our physiology – hair, skin, muscle, eye parts e.g. lenses, etc. They also make up the antibodies for our immune system, hormones, haemoglobin and enzymes.
Proteins are continuously constructed in our cells by enzymes that read and decode (express) genes stored in our DNA. A gene is a section of genetic code which specifies the sequence of amino acids for the protein/enzyme the gene is coding for.
Enzymes
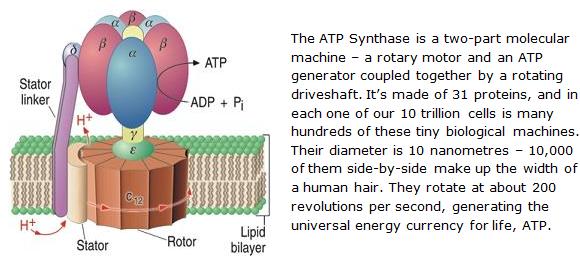
Enzymes are functional proteins. They are catalysts – they speed up the rate of biochemical reactions, carrying out a specific function by interacting with other molecules. Most biological reactions occur very quickly, thanks to the help of enzymes. They are also the biological molecular machines, intricately designed with moving parts, handling and repairing DNA and other tasks. They also perform other functions inside cells such as generating the energy molecule ATP, and transporting molecules around the cell, etc.
An example of an enzyme is the ATP Synthase:
The Genetic Code
The genetic code is the name of the language given to the set of instructions contained in every organism’s DNA. A good comparison is assembly language, which is the low-level programming language used in computers that represents machine code instructions.
The computer’s CPU (central processing unit) interprets data (code) as instructions, according to the CPU’s instruction set. Segments of bits (E.g. 8 bits (byte) or 16 bits (word)) represent the instructions.
In the same way, biological cells interpret the data stored in DNA as instructions, according to the cell’s instruction set, called the Codon Table. Codons are 3 bases long (see drawing below) and could be described as 3-bit bytes (but since each “bit” can be four values, it’s twice binary – quaternary).
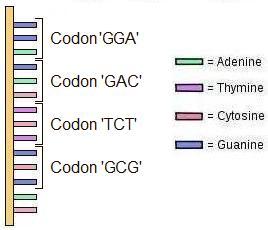
Since each position (base) can be four values (A/C/G/T), each codon can have a value from 0 to 63 (4x4x4). These codons, like computer machine code instructions, virtually index into a lookup table which defines the codon’s instruction. There isn’t a physical look-up table inside a cell, it’s a kind of formalism (it’s not physically anywhere; the values are assigned by certain molecules (tRNAs) which are themselves constructed from the information in their own genes (a vicious circle), which I’ll describe in The Cell section below).
DNA Codon Table
How the codons are assigned. Amino acids are abbreviated e.g. ser=Serine, val=Valine.
TTT - Phe TCT - Ser TAT -Tyr TGT - CysTTC - Phe TCC - Ser TAC -Tyr TGC - CysTTA - Leu TCA - Ser TAA -STOP TGA - STOPTTG - Leu TCG - Ser TAG -STOP TGG - TrpCTT - Leu CCT - Pro CAT - His CGT - ArgCTC - Leu CCC - Pro CAC - His CGC - ArgCTA - Leu CCA - Pro CAA - Gln CGA - ArgCTG - Leu CCG - Pro CAG - Gln CGG - ArgATT - Ile ACT - Thr AAT - Asn AGT - SerATC - Ile ACC - Thr AAC - Asn AGC - SerATA - Ile ACA - Thr AAA - Lys AGA - ArgATG –START* ACG - Thr AAG - Lys AGG - ArgGTT - Val GCT - Ala GAT - Asp GGT - GlyGTC - Val GCC - Ala GAC - Asp GGC - GlyGTA - Val GCA - Ala GAA - Glu GGA - GlyGTG - Val GCG - Ala GAG - Glu GGG – Gly*The START codon doubles with the amino acid Methionine (met).
The Codon Table (above) is the genetic code’s instruction set. There have been many studies into its configuration, which have all drawn the same conclusions – the Codon Table had to be optimally designed, at the origin of life.
These recent studies have proven that the Codon Table’s configuration is finely-tuned for embedded genes (explained shortly) and error-preventing. If the DNA has been damaged by mutations, and the various error correcting systems didn’t find and correct them, the resulting incorrect codes can be very detrimental. The protein being made will end up with the wrong amino acids, which results in reduced or lost function, or worse, destructive. There’s redundancy in the form of multiple codons coding for the same amino acid, and this redundancy appears to be well thought out rather than random. It can easily be seen that most amino acids can handle the last letter changed (e.g. Val can have codes GT?) but it goes further – it’s designed so that if the resulting code does change the amino acid, the incorrect amino acid will be one with similar chemical and physical properties as the correct one, reducing the chances of a negative effect.
The total number of possible combinations of the Codon Table is calculated using the values: 64 (codon count) and 21 (20 amino acids + 1 stop code) = 21^64 = 1.5 x 10^84 which is a 15 with 83 zeros after it. This is around a million times the estimated number of atoms in the universe.
The naturalistic explanation would of course be that it evolved into the optimal configuration over time. This is impossible for two reasons: 1) Any change in codon assignment would change every protein’s configuration made by the cell. Large numbers of proteins would instantly become defective, which would be lethal to the cell. It’s analogous to swapping two letters in the character set of a document – every word containing those two letters would instantly corrupt. For this reason, the Codon Table is naturally locked, because any changes will result in the early death of the organism. 2) Even if the Codon Table could change gradually over time, there isn’t enough time in the history of the universe to test the unimaginable amount of configurations, even if most of the configurations haven’t been tested. For a 14 billion year old universe, 10^54 configurations per second would need to be tested to cover them all.
The Codon Table is precisely configured to avoid decoding errors which are devastating to life. It cannot change without fatal consequences, so evolution is not the answer. It was designed at the beginning by the creator of life.
Genes
Genes code for proteins and enzymes (which as you’ll recall, are made of long chains of amino acids joined together). Basically, genes are packages of information - stretches of codons as described above. Each codon, with the exception of four control codons, encodes for one of the 20 amino acids as shown in the codon table above. Since there are 60 codons coding for 20 amino acids, there is some redundancy – 2-6 codons are assigned to each amino acid which is for error prevention, as described above.
Since the DNA strand is continuous, genes need a way to tell the decoding machinery inside cells where they begin and end. This is accomplished by using the four special control codes. The beginning of a gene is signalled with a Start codon (ATG), and the end of a gene is signalled using one of the three Stop codons (TAA, TAG, TGA).
Using the computer analogy again, genes can be equated to inline subroutines, and cells really are programmable protein makers. Virtually any cell can produce any protein depending on the information it is given, in the form of genes.
All cells carry a full copy of the genome, but certain cells only need certain genes in the genome – for e.g. a liver cell doesn’t need the code for neurons, skin cells don’t need the code for blood components, kidney cells not for hair proteins etc, so – there’s the equivalent of the programming statements “#if” and “#endif” throughout the genome, tailoring it to the type of cell it’s in. This is called Differentiation – which specializes cells to what they’re for – usually while the zygote changes in the womb, but also in later life too. Stem cells basically haven’t yet been given these #if / #endif statements in their copy of the genome to switch off genes – hence stem cells can be used for almost any cell type in surgery.
The Cell
Since this document is predominantly about DNA and the genetic code, and the fact that whole books are needed to cover just the basics, this is only a brief overview of the inner workings of the cell. I’ll mainly focus on how the genetic code is physically read and decoded to make proteins, and the proofreading and error correcting mechanisms cells comprise of which try to keep the genetic code as error (mutation) free as possible.
The human body contains about 100 trillion cells. To realize this number, an average sized classroom filled with sand to the roof has 1 trillion grains of sand. Cells vary quite a lot in size, but the average diameter is around 10 micrometres – 10 of them side-by-side make up the width of a human hair. Human cells generally take about 24 hours to divide, although some take much longer or shorter times, depending their type and purpose.
Humans contain eukaryotic cells, which contain compartments enclosed by membranes in which specific metabolic activities take place. One of the most important compartments is the nucleus – the compartment that houses the cell’s DNA.
Protein Synthesis
The process of synthesizing (making) proteins is called Gene Expression. It generally consists of two major steps: Transcription (an RNA copy is made of the gene from the DNA) and Translation (the codons are decoded into the amino acids which make up the protein).
Transcription occurs within the cell’s nucleus. The enzyme RNA Polymerase, a biological molecular machine, travels down the DNA strand “unzipping” it temporarily so that one of the DNA strands can be copied. When a START codon is found, it begins copying. It produces a single-stranded copy of the DNA’s codes called the pre-messenger-RNA (pre-mRNA), until it finds a STOP codon, which terminates copying. The codes are complementary and T’s are replaced with U’s. This strand undergoes some processing involving cutting and splicing called Alternate Splicing, by more molecular machines - enzymes called Spliceosomes, resulting in a strand called mRNA, which leaves the nucleus into the cytoplasm (the common area of the cell).
Translation is the next step. The mRNA strand is fed into another molecular machine called a Ribosome, which handles the decoding of the codons and formation of the protein’s chain of amino acids. Molecules called Transfer RNA (tRNA) are used to decode the codons into amino acids. These molecules are configured as per the Codon Table. At one end of the molecule is an amino acid and at the other end are three bases which form the complementary codon for that particular amino acid. See diagram below, from Wikipedia:
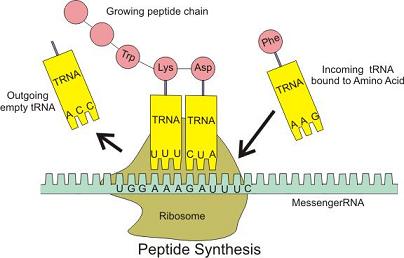
Chains of amino acids are called Peptides. In the diagram above, the mRNA strand is entering the Ribosome from the right hand side, ratcheting in three bases at a time, and the tRNA molecules shuffle until a tRNA’s complementary codon (anticodon) matches the codon on the mRNA strand (the base pairing rules operate here). When one does match, it slots in and the amino acid is attached to the growing chain above.
The rate of translation is quite fast given that the majority of the time is probably taken up ejecting incorrect tRNAs - up to 9 amino acids per second.
When a Stop codon is encountered, translation ceases and the chain of amino acids floats off to another molecular machine called a Chaperonin where it is folded into the desired 3D shape of the protein.
The tRNA Molecules: As just described, decoding the codons is performed mechanically, by tRNA molecules effectively performing bit-testing and logical AND operations on the mRNA strand’s codons. This is an incredible, computer-like process. What’s more, the way the tRNA molecules themselves are made is mindboggling:
There are tRNA genes - their genes are stored in DNA with everything else of course, but are transcribed via another molecular machine called RNA Polymerase III inside the cell’s nucleus. New tRNAs undergo extensive modifications including the attaching of their assigned amino acid (the process is called aminoacylation and involves molecular machines called Aminoacyl tRNA Synthetase) and the attaching of their anticodon.
Therefore, the Codon Table only exists in the code of the tRNA’s genes. It’s amazing that the genetic code’s instruction set is stored in this way, and it’s very difficult to imagine how this level of complex interdependency could have arisen naturally, as it must have corresponded with the origin of life.
Another difficulty for origins is this – the molecular machines I mentioned above are just a few of the hundreds of kinds that occupy the cell, all with their own specific functions. They are enzymes – functional proteins – and have their own genes. They themselves are made via the same process as all the other proteins. I.e. they decode and manufacture themselves, posing a huge problem for naturalistic origins. It’s kind of like having a ‘C’ compiler source code written in C but no executable.
For example, the instructions to build the enzyme RNA Polymerase are themselves encoded in the DNA. But the DNA could not be transcribed into the mRNA without the elaborate machinery of RNA Polymerase. A chicken-and-egg vicious circle, which can be easily solved by the (uncomfortable for some) creation of DNA, genetic code, enzymes, tRNA genes and tRNA molecules all at once.
DNA Proofreading and Error Correcting
DNA’s information is under constant attack from mutations (copying errors) and physical damage. From Wikipedia:
“DNA damage, due to environmental factors and normal metabolic processes inside the cell, occurs at a rate of 1,000 to 1,000,000 molecular lesions per cell per day. While this constitutes only 0.000165% of the human genome's approximately 6 billion bases (3 billion base pairs), unrepaired lesions in critical genes (such as tumour suppressor genes) can impede a cell's ability to carry out its function and appreciably increase the likelihood of tumour formation.”
Sources of damage include: Replicating errors (copying mistakes when cells divide), UV radiation from the sun, cigarettes, x-rays and gamma rays, viruses, and toxic carcinogens to name a few. When nucleobases are changed or deleted the genetic code is corrupted, and this kind of damage is called mutations.
If left unchecked, the rapid genetic meltdown of the organism is certain. Obviously, cells must have the capability to find and repair DNA damage and mutations - and they do: proofreading and error correcting enzymes. These enzymes, like the ones I’ve already mentioned, are molecular machines made of proteins, which all have their building instructions contained in their own genes, stored in the DNA they are required to protect.
Many mechanisms are being discovered, here’s a few:
DNA Polymerase: The enzyme DNA Polymerase is responsible for DNA replication when cells divide. The DNA double strand is "unzipped" and these enzymes run down each unzipped strand, applying the complement bases to form the other side, resulting in two complete and identical DNA double strand structures. At the same time, they proofread and repair the new strand they’re building. After attaching each new base, it checks if that base belongs there according to the base pairing rules, and if an incorrect match is detected, it reverses its direction by one base, that base is cut out and discarded, and the correct base is attached, then it carries on.
This correction mechanism increases the accuracy 100 to 1,000 times. Still, some errors do slip through (and there is the environmental damage to deal with as well) so there are many more mechanisms working to keep DNA as original as possible.
DNA Mismatch Repair: The mismatch repair mechanism is a system for fixing errors that were missed by the DNA Polymerase while replicating DNA. Here is a simplified explanation of this mechanism working in bacteria. Humans have this mechanism as well and is similar, but it’s currently less understood. Three complex enzymes go over the newly copied DNA sequence. The first enzyme senses a distortion in the helix shape of the new DNA, which is caused by incorrectly paired nucleobases. It binds to this region of the DNA strand and signals a second enzyme to come and help with the repair. This second enzyme brings a third enzyme over and attaches the two. Then the third enzyme physically cuts the incorrect base on both sides, and then all three enzymes tag the incorrect section with a methyl group.
At the same time, a new, partial strand of DNA is being created to replace the region in error. Another set of enzymes then cut out the exact amount of DNA needed to fill the gap. Since both the old and new pieces of DNA are present, yet another enzyme decides which one is the new piece, by way of the methyl tag, and then finally this new, correct section is brought over and added to the original DNA strand.
Quite a process involving many molecular machines, all created by the cell following the instructions in their genes which describes their design. They all work together for the sole purpose of keeping errors out of the genetic code and DNA as original as possible.
Broken DNA Strand Repair: Sometimes, the DNA double helix gets broken – both strands are accidently cut. Obviously, this is a huge problem for cells as they cannot cope with such damaged DNA, and this type of damage is a known cause of cancer.
The following mechanism is used to repair broken DNA strands, using enzymes that work to correctly join the DNA strands back together.
Two enzymes work together – one enzyme connects to one of the broken DNA ends and goes searching for other DNA strands that are also disconnected. It attaches itself to all the strands it can find, and studies them to find the one that has the exact nucleobase sequence that’s needed to replace the broken DNA. If this fails, it will go to our second copy of the chromosome to match the sequence for the broken end.
When a correct sequence is found, the enzyme binds the two ends and waits for the second enzyme to help attach them.
Again, complex enzymes that have their structural design encoded in their own genes, which cells use to manufacture all of these tiny molecular machines, whose sole purpose in the universe is to keep the genetic code in DNA as intact and original as possible.
Origins? Life would be a mess without these DNA repair mechanisms – in fact it is very difficult to imagine that life could exist without DNA repair mechanisms. Which poses huge problems for evolution: How did early life cope without them? Can copying errors really be the source of the enzyme’s precise genetic code used to construct them? Supposing early life did survive millions of years without having repair mechanisms, how did so many organisms evolve the same repair systems? The only logical solution is that these mechanisms had to be in place from the very beginning, meaning their coded blueprints were written by the creator of life.
Intelligent Programming?
If what you’ve just read so far sounds incredible, what you’re about to read will dwarf all of that, and could possible cause a paradigm shift in your way of thinking. It’s our DNA and genome we’re talking about here, after all.
If you’re a programmer reading this, the following will be easy to follow and understand. If you’re not a programmer, I hope I have written it in a way for you to at least have a very good idea of the incredible features only being discovered now in our DNA.
Our genetic code appears, in no uncertain terms, to have been intelligently, purposefully, and precisely programmed - the hallmarks of man-made programming techniques. Here’s why:
The human body is estimated to produce over 100,000 different proteins. Since we know that genes tell cells how to make proteins, it was assumed that our genome must therefore contain over 100,000 genes. But after sequencing the human genome and spending years analysing the data, it’s now estimated that our DNA contains roughly 21,000 genes. Obviously, it’s not as straightforward as once thought, and the latest research has discovered a system which allows one gene to produce many different proteins – a process called Alternative Splicing. Added to this, there are two further methods of packing data in DNA – Multiple Bit Streams and Nested/Overlapping Genes, both described after I cover this first one:
Alternative Splicing - LZW Compression used in DNA
Alternative Splicing can be equated to a lossless data compression algorithm very similar to LZW (programmers will be aware of this, others may know the tools that use this algorithm - mainly WinZip and PKZip). LZW is a dictionary-based compression algorithm; segments of data that are used often throughout the data being compressed are held in a “dictionary” so that instead of repeatedly including these segments over and over, only a small code which corresponds to the larger segment in the dictionary needs to be stored.
Before explaining alternative splicing, a feature with genes needs to be explained. Genes are not solid stretches of code from the Start codon to the Stop Codon. They are made up of sections called Exons and Introns. Exons are segments of codons which code for amino acids and Introns are segments of non-coding data (initially labelled “junk DNA”, but the latest research has found that Introns are far from “junk”). In the process of making a protein from a gene, initially a copy strand is made from the DNA (this floating copy is called pre-messenger-RNA or pre-mRNA). Before it can be decoded into the chain of amino acids, the Introns need to be physically cut out. Enzymes – actual biological molecular machines - cut them out and splice the Exons together forming a strand of continuous codons called mRNA. This mRNA strand is then decoded into amino acids for the required protein, as described in The Cell section above.
The data decompression is handled at the splicing stage. Different exons can be spliced together to code for different proteins. See diagram below, from Wikipedia:
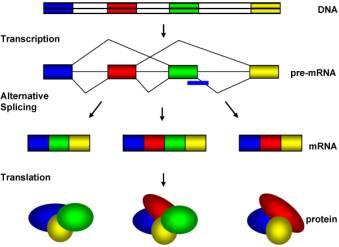
As you can see in this example, three different proteins are produced from the same stretch of DNA. Each coloured section is an exon and could be 100’s to 1,000’s of codons long. The scale of the compression is obvious – in the above example the blue exon could be a couple of thousand nucleobases long and is used in many proteins, but only one copy is stored in DNA. This is how 21,000 genes can code or over 100,000 proteins.
I should add here that the above example shows one gene being used to produce multiple proteins. Latest research has found that exons don't even have to be in the same gene - exons from different genes can be cut out and spliced together to form new proteins. The complexity is staggering.
A great example for showing how this compression is used is in the cochlea (hearing organ) of chickens. It’s lined with hair cells, and each of these cells responds to a specific frequency. One end of the cochlea contains cells that respond to low-frequency sounds and the other end has cells that respond to high-frequency sounds. In between, there are 574 cells that respond to different frequencies, graded from the low end to the high end. The cells respond to different frequencies because of slightly different proteins each one produces. It turns out that a single gene uses alternative splicing to create these 576 different proteins!
Who would have thought that DNA - the fabric of life – uses a lossless data compression algorithm similar to Lempel–Ziv–Welch (LZW)?
Multiple (six) Bit Streams
As you’ll recall, DNA is comprised of two strands. Looking at one strand, there is a stream of bases representing the genetic code. Then there is the adjacent strand with the complementary bases, following the pairing rule: A-T and C-G. It turns out that both strands are read by cells when decoding genes into proteins. This is quite remarkable when you think about it, as the data in the two strands are depended on each other; you can’t change one side without changing the other side, and sometimes there are exons on both sides of the DNA sharing the same base pairs. On top of this, the strands are read in OPPOSITE directions. The two strands are described as antiparallel – their molecules are configured in opposite directions, which means the DNA Polymerase enzymes read the strands in opposite directions to each other.
Added to the above, because codons are 3 bases long, and the cell can start reading at any base, there are three reading frames. Consider the sample code:
GGAGACTCTGCGAC
Depending on which letter you start reading from, the codons are completely different:
GGA GAC TCT GCG AC... or
G GAG ACT CTG CGA C... or
GG AGA CTC TGC GAC...
As you can see, there are 3 possible streams of codons depending on which letter you start from. These are called Reading Frames.
So since both strands are read by the cell’s decoding machinery, and both have three possible reading frames, DNA has six different streams of data. This leads to the next discovery which is difficult to comprehend – genes can be nested inside other genes – the same nucleobases used for more than one gene, on opposite strands and/or reading frames.
Nested / Overlapping Genes
Due to multiple reading frames as described above, it’s easy to understand how the same nucleobases can encode different amino acid sequences, aka genes. It allows two or more genes which encode for entirely different proteins to share the same nucleobase sequence.
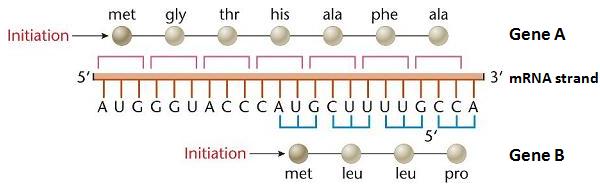
In the above example, the mRNA strand is drawn in orange. Remember, this is the messenger-RNA (mRNA) strand which was copied off DNA at transcription, so “U” replaces “T” plus the bases are inverted (as was described in The Cell above). The 3-letter abbreviations (‘met’, ‘gly’, ‘thr’, etc) are the abbreviations for the amino acids the codons code for. Gene A is indicated across the top, which begins at the start of this strand at the Start Codon (which doubles for the amino acid ‘met’) AUG. Gene B begins 10 bases down the strand indicated by its AUG Start codon, which is frame-shifted right by one base. Both genes use the same bases but code for completely different proteins due to the shifted reading frame.
Genes can overlap as in the above example, as well as be completely embedded within another gene – the embedded gene starts and ends within the host gene. They can share exons, introns, and exist on the opposite (complementary) strand, where the embedded genes are read in the OPPOSITE direction to the host gene. The genes on opposite sides of the strand effectively share bases, due to the base pairing rules.
Thousands of overlapping and embedded genes have been found in the human genome.
Here are a couple of DNA examples, first showing the “conventional” belief of how genes existed in DNA before embedded and overlapping genes were discovered, below it.
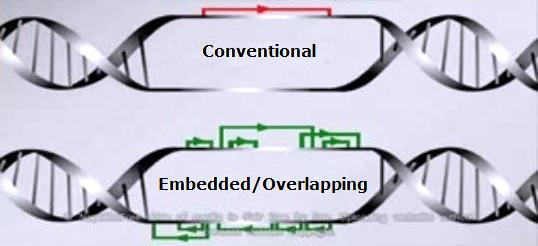
The second diagram above indicates how genes have been found – on their own, overlapping, and completely embedded.
It’s hard to comprehend how multiple overlapping codes were formed. They work in conjunction with each other, rather than interfering with each other. Even more remarkable is the finding of a new study which found that the genetic code is also optimized to harbour the overlapping codes. It seems that the order of the codon table has been tweaked to allow for overlapping genes, as well as for error preventing as already described.
A good analogy is the alphabet. If you wanted to write sentences inside other sentences using the same letters, it would make it easier if the alphabet was designed with this in mind to help make the letters match. As it is, the alphabet wasn’t designed or optimized with this in mind so it’s very difficult to do. Single words can work - for e.g. “thunder” is within the sentence “They’re both under the weather”, but whole sentences can be next to impossible. Yet, overlapping and embedded genes are quite common. If this wasn’t enough, the genes that are written BACKWARDS within other genes defy understanding, just as sentences appearing backwards in paragraphs would be.
For the programmers reading this, it would be similar to programming a collision system, and at the same time making sure that its resulting machine code could be executed BACKWARDS for your high-score routines. It really is ingenious.
Summary
Twelve pages and I haven’t even scratched the surface. This document is only intended to give the reader a foundational knowledge of DNA and genetics, the genetic code, plus a basic understanding of gene expression into proteins. Here’s a brief recap:
- DNA is an information storage molecule. It stores digital data in quaternary (twice binary) so densely that a pinhead of DNA equals 2 million 2-terabyte hard drives, and the length of this double helix structure would circle the earth 30 times.
- The base-pairing design of DNA allows it to be easily replicated, error-checked, and repaired.
- The human genome is about 3 billion base pairs long, making the DNA structure around 6 feet long, and contains the equivalent of 750 Mbytes – a CD’s capacity.
- Proteins are made from long chains of 20 different amino acids, which are the building blocks of life. The genetic code in genes tells cells how to make each of the 100,000+ different proteins in the human body. Genes can be described as protein recipes, as they list the amino acids required and their configuration.
- Structural proteins include hair, nails, skin, muscle, bones, eye ball parts, etc.
- Functional proteins, called enzymes, have the ability to carry out metabolic processes. It’s accurate to call them biological molecular machines because that is how they are designed and is how they function. Many have multiple moving parts to carry out their work.
- The genetic code is the name of the language given to the set of instructions contained in every organism’s DNA, and is comparable to computer assembly language.
- The Codon Table is the genetic code’s instruction set. Each instruction is called a codon and is 3 bases long and each has 64 possible values. Codons code for amino acids plus the control codes Start and Stop.
- The Codon Table is finely-tuned to avoid decoding errors and to allow for embedded genes. It couldn't have evolved because any changes have fatal consequences. It had to be present in its finely-tuned configuration right from the beginning – at the origin of life.
- Genes are packages of information - stretches of codons which code for proteins and enzymes. They begin with a Start codon and end with a Stop codon. They are analogous with computer inline subroutines. Because they code for proteins, cells really are programmable protein makers – software driven.
- A main function of cells is gene expression – to synthesise proteins according to the instructions contained within genes. First the gene is transcribed off DNA onto an mRNA strand, then this is cut and spiced, then it’s translated into a chain of amino acids, finally the chain is folded into a 3D shape.
- Alternative Splicing can be equated to a lossless data compression algorithm very similar to Lempel–Ziv–Welch (LZW), which allows 21,000 genes to code for 100,000+ proteins.
- To considerably increase DNA’s storage capacity, there are six reading frames – three each side of the strand. Three bit streams using the same nucleobases. The Codon Table has been finely-tuned to allow for this.
- For storing even more data in DNA there are embedded genes. Genes written within genes, sharing the same nucleobases. They can overlap, be totally embedded within the host gene, even being read in the OPPOSITE direction to the host gene. Again, the Codon Table has been finely-tuned to allow for this as well.
Conclusion
Many people have a false belief about genetics - that DNA is “just” chemicals, whose molecular structure merely interacts with other molecules and the resulting proteins are only produced because of the “patterns” in the DNA. This is clearly not correct. As I have shown, the genetic code is digital (quaternary) and is written using the codes from the Codon Table, which in itself proves it’s a coded language. Genes are akin to inline subroutines in programming, marked out in the genome by Start and Stop codes. Cells are programmable protein makers – the same biological machinery makes almost any protein simply by reading and decoding the instructional information held in genes. Gene expression is clearly not a chemical reaction, but digital information processing.
Genes contain instructional information which only comes from intelligence. The cell’s biological systems are clearly information-based. What’s more, the information is structured into a language and is highly organized just like you would find in the source code of programmers developing data-intensive applications. This requires fore-thought and planning, which can be seen when you really examine the genetic code closely. The hallmarks of a pure genius at work are clearly visible. I’ve only just scratched the surface of the “genetic operating system” and discovered multiple parallel bit streams, overlapping code, and a sophisticated data compression algorithm that allows over a 100,000 proteins to be coded from just 21,000 genes. Plus, the integrity of this system (and therefore our lives) is protected by multiple layers of proofreading and error correcting mechanisms which increase DNA’s accuracy by thousands of times – again, a feature that is crucial in man-made data communications over the internet, telecommunications and data storage. Finally, the genetic code’s instruction set, the Codon Table, has been finely-tuned for two crucial reasons: To allow for embedded and overlapping genes, and for preventing detrimental errors in translation.
The naturalistic explanations for the origin of life and the diversity of it is abiogenesis (life from non-living chemicals) followed by evolution.
Abiogenesis: In 1953 scientists Stanley Miller and Harold Urey performed a now-famous origins-of-life experiment (the Miller-Urey Experiment). They had to assume that the early Earth had no oxygen in the atmosphere, because amino acids are destroyed by oxygen. But if their assumption is correct, then there would not have been any ozone in the atmosphere either. Ozone protects us from the sun’s UV rays for many good reasons; one is that nucleic acids are rapidly decomposed by UV light - nucleic acids are essential for life and include DNA and RNA, so trying to solve one problem just causes another. They did produce some amino acids, but amino acids are far from “life”. “Organic” compounds like amino acids are molecules containing carbon - it doesn’t mean much more than that. In any case, geochemists now conclude that Earth’s early atmosphere was nothing like the mixture of gasses used in this experiment, and that the experiment would not have produced anything meaningful for life with the gases they believe were present in an early-Earth atmosphere.
Amino acids and other organic compounds are found throughout the universe, on meteorites and therefore planets such as Mars, are as far from life as alphabet letters are from computer software code. Life is informational and without information and the biological molecular machines to read, interpret, and manufacture everything, life isn’t possible. Not to mention the materialist’s belief that life is just chemicals, but that’s for another time.
Lastly, there is no “deep time” for the first cell to have formed naturally. According to Wikipedia, the “Late Heavy Bombardment” ended 3.9 billion years ago. This means that the new Earth cooled and became habitable at that time. But also from Wikipedia, the first organism evolutionists call the Last Universal Common Ancestor (LUCA) (the first organism on Earth that all Earth’s plants and animals descended from) lived “3.5 to 3.8 billion years ago”. This leaves about 200 million years to go from non-living chemicals to cells with the “genetic operating system” that is described in this document, and more. To verify this, Wikipedia lists the features the LUCA had to have, which includes DNA storing genetic code, error correcting mechanisms, and the many other biological molecular machines.
Panspermia? Panspermia is the hypothesis that life exists throughout the Universe, and that bacteria was transferred to the early Earth via comets and asteroids. Apart from the fact that any life would be burnt to a crisp as it entered Earth’s atmosphere (several years ago, an experiment which attached rocks containing bacteria to the heat shield of a returning rocket proved this), this doesn’t explain the origin of life itself.
Evolution: The process that results in heritable changes in a population spread over many generations. Evolutionary processes are responsible for the diversity of life – including species, individual organisms and molecules. Specifically, this is how the Theory of Evolution explains the existence of all life on Earth – after life on Earth originated (abiogenesis), it then evolved from the Last Universal Common Ancestor (LUCA) into all of the plants, animals and humans of today. The LUCA is said to be a small, single-celled organism much like modern bacteria. This then evolved into multicellular life, then into both plants and animals. Staying on the animal branch - they evolved into fish, then into amphibians, and then branched off into reptiles and mammals. Reptiles evolved into birds. Some mammals returned to the sea to become the sea mammals (seals, whales, dolphins, and walruses). Other mammals evolved into all the mammals of today including humans via apes. That was just a very quick overview.
Ignoring the gigantic leap from non-living chemicals to the first organism resembling bacteria (where the digital format, Codon Table, and enzymes all had to come into existence), bacteria have relatively small genomes at a few hundred thousand base pairs. That’s 0.01% the size of a human’s genome of 3 billion base pairs. As previously mentioned, the human genome is contained in a 6 foot long DNA strand. Six feet = 2 metres = 2,000 millimetres. 0.01% of 2,000 mm = 0.2 mm. So the length of a bacteria’s genome is around a quarter of a millimetre - as long as the width of two strands of human hair. For bacteria to evolve into humans, the genome has to increase from a quarter of a millimetre to six feet long, filling the DNA with thousands of genes coding for over a hundred thousand proteins.
Therefore, for evolution to work, genomes need to increase in size and new information added. The only source of new genetic code is from mutations – copying errors that change the nucleobases – and the belief is that natural selection will keep the “best” genetic sequences, generation to generation. Everything that organisms have today that bacteria do not have – blood, hearts, livers, eyes, brains, etc – the genes which code for all of these parts, and all of the controls and other genetic information, had to come from copying mistakes.
One example from thousands - multiple genes totalling thousands of nucleobases make haemoglobin proteins in blood. Under the evolutionary idea, these genes had to “error-up” into existence to form blood. But anything less than these genes is not blood, and there cannot be any natural selection on non-functioning genes.
Apart from these glaringly-obvious problems for evolution, here is a summary of the other main points which the theory of evolution fails on:
- Genes code for enzymes, the cell’s biological molecular machines, but enzymes are required to decode the genes. A vicious circle for origins – they decode themselves. So both their genes and themselves need to originate together, at the beginning of life.
- Proofreading and error correcting mechanisms – these complex enzymes and DNA repair systems needed to be present and functional at the beginning of life.
- Alternative Splicing, the LZW data compression algorithm – this is an intelligent system that cannot evolve from copying errors.
- Bit streams / embedded genes – again, a complicated system which cannot evolve from copying errors.
- The genetic code’s instruction set (the Codon Table) is finely-tuned. There are more possible configurations than there are atoms in the universe, and the configuration chosen is virtually the optimal one for resisting errors caused by mutations, resisting errors during translation, and allowing overlapping and embedded genes. It was configured right at the beginning of life as it is impossible for it to change/evolve. Also, it was the current configuration right at the beginning of life because most all of life share this same configuration.
- Obviously, there are evolutionary ideas as to how the genetic code evolved into the current configuration, but none are conclusive and most offer “just-so” stories including Convergent Evolution – the fact that identical biochemical systems, the same nucleobase sequences, the same amino acid sequences, the same 3D structures can be found in organisms who are not directly related to each other, forces the evolutionist to conclude that the same systems evolved independently. Highly improbable, since they’re all based on random mutations.
- Another study notes that the Codon Table is not the best configuration – but they’re only testing for error-prevention and not including the trade-off needed for overlapping/embedded genes – making the Codon Table “best for both”.
The cell’s information systems and processes are strikingly similar to man-made ones. Design is easily recognisable and is understandable as most of the processes follow similar pathways as our own information systems.
Each one of the cell’s components is enormously complex and necessary to its functioning and replication, but each component alone cannot survive nor replicate itself. Until everything works, nothing works. There are no possible paths for evolution to take to build the systems contained in biological cells.
If scientists are only allowed "natural" explanations, then they are not pursuing the truth; they are pursuing a natural explanation - even if a natural explanation is impossible.
The ink and paper did not write the book. The evidence is clear and easily understood – the only reasonable, logical, and possible explanation is that it must have been the work of a Creator, who obviously created us and everything else for a purpose.
References:www.wikipedia.com, www.nature.com, www.sciencedaily.com, www.ncbi.nlm.nih.gov, www.pnas.org, www.rcsb.org, www.uniprot.org, www.chemguide.co.uk, www.biotech.about.com, www.atpsynthase.info